Aetiology
Virology
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a previously unknown betacoronavirus that was discovered in bronchoalveolar lavage samples taken from clusters of patients who presented with pneumonia of unknown cause in Wuhan City, Hubei Province, China, in December 2019.[26]
Coronaviruses are a large family of enveloped RNA viruses, some of which cause illness in people (e.g., common cold, severe acute respiratory syndrome [SARS], Middle East respiratory syndrome [MERS]), and others that circulate among mammals and birds. Rarely, animal coronaviruses can spread to humans and subsequently spread between people, as was the case with SARS and MERS.
SARS-CoV-2 belongs to the Sarbecovirus subgenus of the Coronaviridae family, and is the seventh coronavirus known to infect humans. The virus has been found to be similar to SARS-like coronaviruses from bats, but it is distinct from SARS-CoV and MERS-CoV.[27][28]
See the Classification section below for information on SARS-CoV-2 variants.
[Figure caption and citation for the preceding image starts]: Illustration revealing ultrastructural morphology exhibited by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) when viewed with electron microscopicallyCenters for Disease Control and Prevention [Citation ends].
Origin of virus
A majority of patients in the initial stages of this outbreak reported a link to the Huanan South China Seafood Market, a live animal or ‘wet’ market, suggesting a zoonotic origin of the virus.[29][30][31] An initial assessment of the transmission dynamics in the first 425 confirmed cases found that 55% of cases before 1 January 2020 were linked to the market, whereas only 8.6% of cases after this date were linked to the market. This suggests that person-to-person spread was occurring among close contacts since the middle of December 2019.[31] Other studies suggest that the virus may have emerged earlier than previously thought in other countries.[32][33]
A zoonotic origin has not been confirmed. Some studies suggested that SARS-CoV-2 may be a recombinant virus between a bat coronavirus and an origin-unknown coronavirus, with pangolins and minks suggested as possible intermediate hosts. However, there is currently no evidence to demonstrate the possible route of transmission from a bat reservoir to humans through one or several intermediary animal species.[34]
An independent assessment of the origins of SARS-CoV-2 concluded that there are data gaps that preclude determining how the virus initially entered the human population with any certainty.[35]
Transmission dynamics
Respiratory transmission is the dominant mode of transmission, with proximity and ventilation being the key determinants of transmission risk.[36] Available evidence suggests that transmission between people occurs primarily when an infected person is in close contact with another person. The virus can spread from an infected person’s mouth or nose in small liquid particles (ranging in size from larger droplets to smaller aerosols) when the person coughs, sneezes, sings, breathes heavily, or talks. Close-range contact can result in inhalation of, or inoculation with, the virus through the mouth, nose, or eyes.[37]
Aerosol transmission can occur in healthcare settings during aerosol-generating procedures. There are also some outbreak reports that suggest aerosol transmission is possible in the community under certain conditions; however, these reports relate to enclosed indoor crowded spaces with poor ventilation where the infected person may have been breathing heavily (e.g., restaurants, choir practice, fitness classes).[37][38] A detailed investigation of these clusters suggests that droplet and fomite transmission could also explain the transmission in these reports.[37] While the air close to, and distant from, patients has been found to frequently be contaminated with SARS-CoV-2 RNA, few of these samples contained viable virus.[39] The risk of transmission is much lower outdoors compared with indoors, with a limited number of studies estimating a transmission rate of <1%.[40][41] Evidence that nebuliser treatments increase the risk of transmission of coronaviruses similar to SARS-CoV-2 is inconclusive, and there is minimal direct evidence about the risk for transmission of SARS-CoV-2.[42]
Fomite transmission (from direct contact with fomites) may be possible, but there is currently no conclusive evidence for this mode of transmission. In the few cases where fomite transmission has been presumed, respiratory transmission has not been completely excluded.[36] While the majority of studies report identification of the virus on inanimate surfaces, there is a lack of evidence to demonstrate recovery of viable virus.[43] Replication-competent virus is more likely to be identified when polymerase chain reaction cycle threshold for clinical specimens from infected individuals is <30 (i.e., high viral load).[44]
Faecal-oral transmission (or respiratory transmission through aerosolised faeces) may be possible, but there is only limited circumstantial evidence to support this mode of transmission.[36]
Transmission via other body fluids (including sexual transmission or bloodborne transmission) has not been reported.[36][45] While the virus has been detected in body fluids (e.g., semen, urine, cerebrospinal fluid, ocular fluids), the presence of virus or viral components does not equate with infectivity.[46] There are limited data about the transmission risk from organ donors. However, there appears to be a low risk of transmission with non-lung (i.e., kidney, liver, heart) organs from SARS-CoV-2-positive donors, irrespective of whether the donor is symptomatic at the time of procurement.[47]
Perinatal (vertical) transmission occurs rarely and transplacental transmission has been documented. There is limited evidence on the extent of vertical transmission and its timing.[48] Further high-quality studies are required to establish whether perinatal transmission occurs.[49] Viral fragments have been detected in breast milk; however, this finding is uncommon and, when it occurs, has been associated with mild symptoms in infants.[50]
Nosocomial transmission was reported in 44% of patients in one systematic review; however, this review was limited to case series conducted early in the outbreak in Wuhan before the institution of appropriate infection prevention and control measures.[51] Hospital-acquired infections accounted for approximately 11.3% of infections in the UK between February and August 2020 (rates as high as 25% were reported). Rates were notably higher in residential community care hospitals (61.9%) and mental health hospitals (67.5%) compared with acute and general care hospitals (9.7%).[52][53] Studies of healthcare workers exposed to index cases (not in the presence of aerosol-generating procedures) found little to no nosocomial transmission when contact and droplet precautions were used.[54] The risk to healthcare workers performing or assisting with a tracheostomy appears to be low.[55]
BMJ: visualising SARS-CoV-2 transmission routes and mitigations Opens in new window
Transmission dynamics in relation to symptoms
Transmission is more likely if contacts are exposed shortly before or after symptom onset in the index patient.
The risk of transmission to close contacts was higher if exposure occurred between -2 and 3 days from symptom onset in the index patient in one study. Among contacts who became infected, asymptomatic infection was more common if they were exposed to an asymptomatic index patient, suggesting that disease severity in the index patient may be associated with the clinical presentation of disease.[56]
The median duration of infectiousness in patients with mild disease in a real-world community setting (pre-Omicron variant) was approximately 5 days (range 3 to 7 days) in one study. Symptom onset was a median of 3 days before peak viral RNA and infectious viral load. Less than 25% of cases shed infectious virus before symptom onset. Two-thirds of cases were still infectious 5 days after symptom onset, and one third were still infectious at 7 days.[57]
Onward transmission varies according to specific host and contact factors, and the nature of the exposure. Factors associated with increased transmission include:[58]
Environmental factors: indoors, poor ventilation, crowding, close proximity, shared facilities, cold ambient temperature, low humidity
Host factors: recently infected, high viral load, severe disease, age, presence of comorbidities, immunocompromised
Behavioural factors: singing/shouting, coughing/sneezing, hugging/kissing, mask etiquette, hand hygiene, duration of contact
Viral factors: changes in the viral genome linked to increased transmissibility.
Symptomatic transmission
Transmission mainly occurs via respiratory droplets or aerosols during close contact with an infected symptomatic case. Transmissibility depends on the amount of viable virus being shed and expelled by a person, the type of contact, the setting, and what infection prevention and control measures are in place.[37]
Presymptomatic transmission
Transmission may occur during the incubation period before symptom onset.
While there is evidence of transmission from presymptomatic people, there is limited evidence on how frequently this is likely to occur and estimated transmission rates are highly variable.[59][60]
Only 7% of people exposed to a presymptomatic index case became infected in one systematic review.[61] After controlled experimental inoculation with SARS-CoV-2 virus, only 7% of virus emissions into the air and environment occurred before the first reported symptom, suggesting that presymptomatic transmission is responsible for a small fraction of infections.[62]
People without symptoms may be presymptomatic, or they may remain persistently asymptomatic.
Asymptomatic transmission
Transmission from asymptomatic cases (laboratory-confirmed cases who never develop symptoms) has been reported; however, most of the evidence is based on early data from China and has limitations (e.g., small number of cases, cases may have been presymptomatic).[63][64][65][66][67][68] Numerous studies have reported no evidence of asymptomatic transmission from carriers of SARS-CoV-2, including a large study in nearly 10 million residents in Wuhan.[69][70][71][72] Only 1% of people exposed to an asymptomatic index case became infected in one systematic review, suggesting limited infectiousness.[61]
Evidence on the transmission dynamics associated with the Omicron variant is mixed, with some studies suggesting that symptomatic cases were more likely to transmit the infection compared with asymptomatic cases, and other studies showing no difference.[73]
Estimating the prevalence of asymptomatic cases in the population is difficult. One living systematic review and meta-analysis found that the interquartile range for the proportion of persistently asymptomatic cases was 14% to 50% across studies; however, heterogeneity was high so the study did not estimate a mean proportion of overall asymptomatic infections.[74] A meta-analysis of over 130,000 people found that 21.7% remained asymptomatic throughout the course of the infection (after excluding presymptomatic cases). Subgroup analysis showed that the overall rate of asymptomatic infections was higher in pregnant women (48.8%) and children (32.1%). African studies reported the highest asymptomatic infection rate, while Asian studies reported the lowest.[75] The pooled percentage of asymptomatic infections has been estimated to be 25.5% to 32.4% among patients infected with the Omicron variant.[76]
Healthcare workers may play a role in asymptomatic transmission. About 7.6% of healthcare workers who worked in hospital units with infected patients tested positive for SARS-CoV-2 antibodies; however, only 58% of these workers reported prior symptoms.[77]
Although there is some evidence that older children have higher rates of asymptomatic disease than infants <1 year of age, the majority of children present with symptomatic disease and do not appear to be silent spreaders of infection.[20]
Superspreading events
Superspreading events are associated with explosive growth early in an outbreak and sustained transmission in later stages. Examples include church/religious gatherings, family or social gatherings, choir practices, indoor recreational sporting activities, nightclubs, restaurants, business conferences, and working in call centres. Widespread transmission has also been reported in long-term care facilities, homeless shelters, prisons, and meat and poultry processing facilities, as well as on board cruise ships.[78]
Limited transmission has been reported in childcare, school, and university settings.[79][80] There is limited high-quality evidence to quantify the extent of transmission in schools, or to compare it with community transmission. However, evidence suggests a lower overall infection attack rate in school staff (1.18%) compared with students (1.66%). Evidence suggests the overall infection attack rate and SARS-CoV-2 positivity rate in school settings are low.[81][82] During periods of low incidence of infection in the local population in schools with non-pharmaceutical interventions in place, the risk to school staff is not generally higher than that of the general population and not comparable to other high-risk professions (e.g., healthcare workers). Studies reporting periods of high incidence of infection are limited, but do show a higher risk to school staff in these circumstances.[83] In one study, infection in close contacts in secondary schools and colleges in England was uncommon (approximately 2%).[84]
Some individuals are supershedders of virus, but the reasons underlying superspreader events are often more complex than just excess virus shedding and can include a variety of behavioural, host, and environmental factors.[85]
Viral transmission factors
Incubation period
The mean incubation period has been estimated to be between 5 and 7 days, and may differ depending on the variant.[86] The mean incubation period has decreased gradually from the wild-type virus (5.2 days) to the Omicron variant (3.42 days). The overall pooled incubation period was 6.57 days (range 1.8 to 18.87 days), and was higher in older adults (7.43 days) and children (8.82 days).[87]
Reproduction number (R₀)
A systematic review and analysis estimated the reproduction number to be 2.69 (based on published literature from January to August 2020). However, the R₀ differs depending on various factors.[88]
Secondary attack rate
The pooled secondary attack rate among all close contacts of an index case has been estimated to be 7%, based on data from early in the pandemic.[89] However, the rate varied between contact settings with an estimated rate of 18.9% to 37.3% in household settings, 42% among residents in aged-care facilities, 3.6% in healthcare facilities, 1.2% to 5.9% in social settings, and 1.9% in workplaces. The rate is higher for symptomatic index cases compared with asymptomatic cases, and adults compared with children.[74][90][91][92][93][94] The rate in children and young people was higher in household settings compared with school settings.[95] The secondary attack rate in household settings due to transmission from a paediatric index case ranged from 0% to 75% across studies.[96] The secondary attack rate for the Omicron variant is higher compared with other SARS-CoV-2 variants.[93][97]
Viral load
Viral load appears to be a leading driver of virus transmission; higher viral loads are associated with increased secondary attack rates and a higher risk of developing symptomatic disease.[98] Viral load is highest in the upper respiratory tract (nasopharynx and oropharynx) early in the course of infection (usually peaks in the first week of illness), and then increases in the lower respiratory tract (sputum). Viral load decreases after symptom onset. Patients with severe disease have higher viral loads compared with those with mild disease. Viral load in the upper respiratory tract is comparable in asymptomatic and symptomatic patients; however, most studies demonstrate faster viral clearance among asymptomatic people compared with symptomatic people.[99]
Viral shedding
The mean duration of viral shedding depends on the specimen: 17 days in the upper respiratory tract (maximum 83 days); 14.6 days in the lower respiratory tract (maximum 59 days); and 17.2 days in stool (maximum 126 days). Duration of shedding was longer in symptomatic patients compared with asymptomatic patients, in children compared with adults, and in patients with severe illness compared with those with non-severe illness.[99][100] Immunocompromised patients may shed for at least 2 months.[101] There are reports of super shedders who shed the virus for prolonged amounts of time (the longest was a case who tested positive for 505 days).[102] There is no convincing evidence that duration of viral shedding correlates with duration of infectivity.[103] No viable virus has been isolated in patients with mild or moderate disease after 10 days of symptoms, or after 20 days in those with severe or critical disease, despite ongoing viral shedding.[36] Fully vaccinated people may have a shorter duration of viable shedding compared with partially vaccinated or unvaccinated people.[104]
The pooled duration of viable virus shedding was 5.2 days for the Omicron variant, with the duration being slightly higher in symptomatic patients compared with asymptomatic patients (although the difference was not significant).[105]
Pathophysiology
The exact pathophysiology is unknown, partly due to the scarcity of postmortem studies.[106] The pathophysiology resembles that of other coronavirus infections. However, some evidence indicates that COVID-19 has distinctive pathophysiological features that set it apart from respiratory failure of other origins.[107]
SARS-CoV-2 attaches to the angiotensin-converting enzyme-2 (ACE2) receptor on target host cells, followed by internalisation and replication of the virus. ACE2 receptors are highly expressed in the upper and lower respiratory tract cells, but are also expressed in myocardial cells, renal epithelial cells, enterocytes, and endothelial cells in multiple organs, which may explain the extrapulmonary manifestations associated with the disease.[108] Viral RNA has been identified in many organs in postmortem studies.[106]
[Figure caption and citation for the preceding image starts]: Multi-organ complications of COVID-19 and long COVID. The SARS-CoV-2 virus gains entry into the cells of multiple organs via the ACE2 receptorBMJ. 2021;374:n1648 [Citation ends].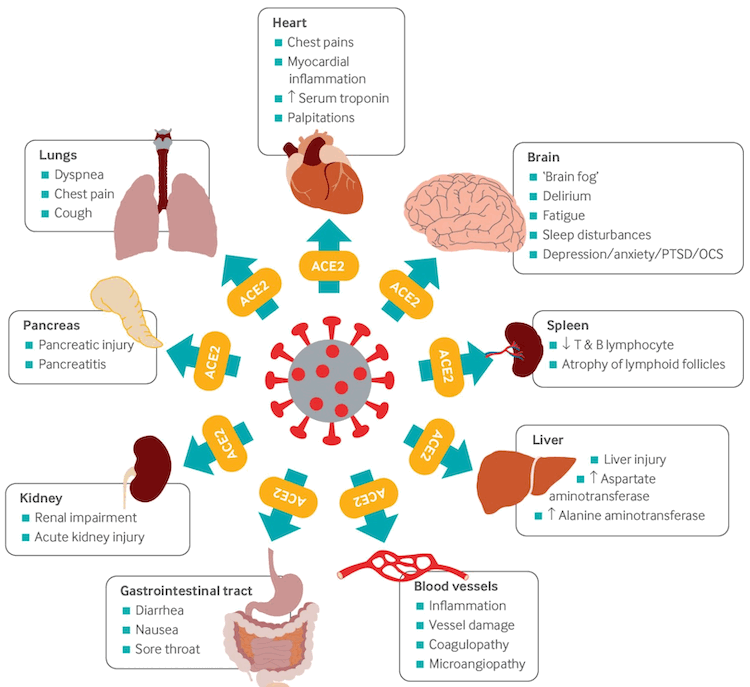
The virus uses host transmembrane protease serine 2 (TMPRSS2) for viral spike protein priming and fusion of viral and host cell membranes.[109] The SARS-CoV-2 spike protein plays a key role in the recognition of the ACE2 receptor and cell membrane fusion process. A unique structural feature of the spike glycoprotein receptor-binding domain confers potentially higher binding affinity for ACE2 on host cells compared with SARS-CoV-1.[110] This furin-like cleavage site does not appear to exist in other coronaviruses.[111] The binding energy between the spike protein and ACE2 was highest for humans out of all species tested in one study, suggesting that the spike protein is uniquely evolved to bind to and infect human cells expressing ACE2.[112] Evidence suggests that the spike protein alone may damage endothelial cells by downregulating ACE2 and consequently inhibiting mitochondrial function. Further research is required on whether the spike protein can by itself trigger cell signalling that could lead to various biological processes.[113][114] SARS-CoV-2 variants may be more transmissible, at least in part, due to enhanced spike protein binding affinity for the ACE2 receptor.[115]
[Figure caption and citation for the preceding image starts]: Virus replication cycleBMJ. 2020;371:m3862 [Citation ends].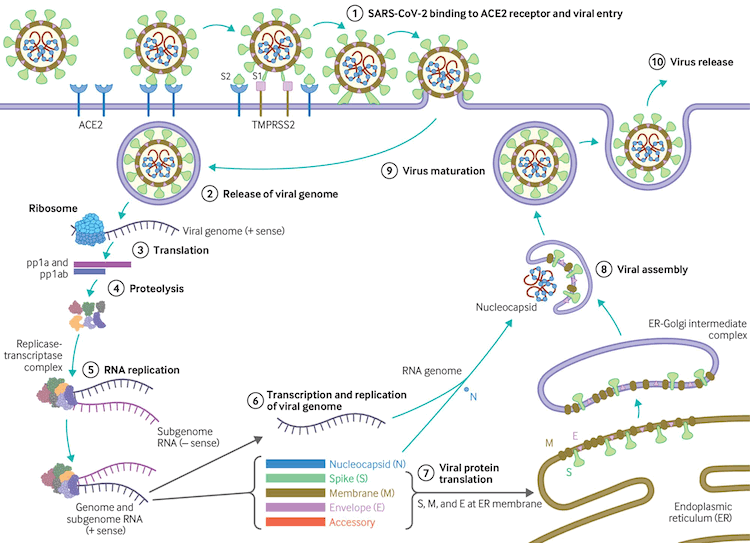
In addition to direct cytopathic viral injury, severe disease is frequently complicated by an infection-induced microangiopathy or hypercoagulable state that causes capillary, venous, and/or arterial thrombosis, which may lead to end-organ damage due to distant thrombotic or embolic disease. Widespread microthrombi have been identified in almost every organ in postmortem studies. The predominant pathological findings in fatal cases were diffuse alveolar damage, coagulopathy, and haemodynamic compromise. Involvement of non-pulmonary organs was limited to mild parenchymal inflammation (e.g., myocarditis, hepatitis, encephalitis). Direct viral cytopathic injury of extrapulmonary organs in general was not regarded as the cause of organ failure.[106][108][116] The majority of findings in non-pulmonary organs were related to chronic diseases.[117] SARS-CoV-2-induced endotheliitis may play a role in both the respiratory and non-respiratory manifestations.[118]
Three major tissue phenotypes have emerged in postmortem lung tissue: a classic phenotype characterised by progressive diffuse alveolar damage; bronchopneumonia from secondary infection; and tissue thrombosis. These phenotypes are not mutually exclusive and may overlap.[119] Severe pulmonary disease is a consequence of fibrotic remodelling and secondary lobular microischaemia, resulting in a distinctive form of fibrotic interstitial lung disease.[120]
SARS-CoV-2 placentitis is a distinct pathological entity that has been reported in pregnant women, and is characterised by massive perivillous fibrin deposition and chronic histiocytic intervillositis. It is associated with increased risk of pregnancy loss.[121]
Genetic factors may play a role in susceptibility to infection and disease severity; however, further research is required.[122][123]
Classification
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant classification
All viruses, including SARS-CoV-2, change over time. Most changes have little to no impact on the virus’ properties; however, some changes may affect virus transmission, disease severity, and performance of diagnostic tests, therapeutics, or vaccines.
SARS-CoV-2 variants have been emerging and circulating around the world since the beginning of the pandemic, and are routinely monitored and classified as either variants under monitoring, variants of interest, or variants of concern by the World Health Organization (WHO).[2] These classification systems may vary between countries.
The WHO assigns simple labels for variants of concern using letters of the Greek alphabet. Previously, Greek labels were also used to name variants of interest, but this is no longer the case. This labelling does not replace existing scientific names (e.g., Pango, Nextstrain, GISAID), which continue to be used in research.[2]
Variant under monitoring
The WHO defines a variant under monitoring as a variant with genetic changes that are suspected to affect virus characteristics and early signals of growth advantage relative to other circulating variants (e.g., growth advantage that can occur globally or in only one WHO region), but for which evidence of phenotypic or epidemiological impact remains unclear, requiring enhanced monitoring and reassessment pending new evidence. If a variant has an unusually large number of mutations in known antigenic sites, but with very few sequences and it is not possible to estimate its relative growth advantage, such a variant can also be designated a variant under monitoring if there is also evidence of community transmission in two or more countries within a 2-4 week period.[3]
Variant of interest
The WHO defines a variant of interest as a variant with genetic changes that are predicted or known to affect virus characteristics such as transmissibility, virulence, antibody evasion, susceptibility to therapeutics and detectability; and identified to have a growth advantage over other circulating variants in more than one WHO region with increasing relative prevalence alongside increasing number of cases over time, or other apparent epidemiological impacts to suggest an emerging risk to global public health.[3]
Variant of concern
The WHO defines a variant of concern as a variant that meets the definition of a variant of interest (see above) and determined to be associated with a moderate or high level of confidence (through a risk assessment performed by WHO), meets at least one of the following criteria when compared with other variants:[3]
Detrimental change in clinical disease severity, or
Change in epidemiology causing substantial impact on the ability of health systems to provide care to patients with COVID-19 or other illnesses and therefore requiring major public health interventions, or
Significant decrease in the effectiveness of available vaccines in protecting against severe disease.
Former variants of concern
SARS-CoV-2 variants of concern are now extinct, with the current circulating variants of interest and variants under monitoring being the various Omicron subvariants (see below). Variants have sequentially replaced each other since the start of the pandemic, with the most successful variants being Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (B.1.1.529). Alpha and Delta emerged in late 2020, with Omicron emerging in late 2021. Beta (B.1.351) and Gamma (P.1) were also variants of concern.[4]
The Alpha variant was more transmissible than the wild-type virus, and was associated with an increased risk of hospitalisation and intensive care unit admission (suggesting more severe disease), but not mortality, compared with the wild-type virus, although data are conflicting. It was not associated with changes in the symptoms reported or their duration.[5][6][7][8]
The Delta variant was more transmissible than the wild-type virus and Alpha variant, and was associated with an increased risk of hospitalisation (suggesting more severe disease) compared with contemporaneous Alpha and Beta cases. However, there was a high level of uncertainty in the findings. The crude case fatality rate was less than the Alpha variant.[9]
Omicron variant
The Omicron variant comprises the parental lineage B.1.1.529 and its descendent lineages (or subvariants) BA.1, BA.2, BA.3, BA.4, and BA.5, as well as their various sublineages. Omicron subvariants are currently in circulation.
The WHO considers the classification of Omicron sublineages independently as variants under monitoring, variants of interest, or variants of concern. Previously, Omicron sublineages were all classified as part of the Omicron variant of concern. This classification did not have the granularity needed to compare new descendent lineages with altered phenotypes to the Omicron parent lineages BA.1, BA.2, BA.3, BA4, and BA.5.[10]
Omicron is a highly divergent variant with a high number of mutations. There is no path of transmission linking Omicron to its predecessors (Alpha, Delta), and it has been estimated that its closest-known genetic ancestor likely dates back to some time after mid-2020.[11] Cases were first reported in South Africa in November 2021. Omicron showed substantial growth advantage over Delta, and rapidly replaced Delta globally. The Omicron variant became the dominant variant in many countries, and accounted for over 98% of publicly available sequences since February 2022.[10]
Evidence suggests reduced disease severity and lower mortality for the Omicron variant compared with the Delta variant, after adjusting for the confounding effects of age, sex, ethnicity, prior infection, vaccination status, comorbidities, and effect of province and effect of public/private sector.[12][13] The majority of deaths occurred in adults aged ≥65 years and patients with ≥3 underlying medical conditions.[14] Evidence from animal studies and ex vivo cultures of the human lower and upper respiratory tract suggests that Omicron does not infect cells deep in the lung as readily as it does cells in the upper airways.[15][16] Transmission, disease severity/hospitalisation rate, and vaccine efficacy may differ depending on the specific Omicron sublineage. Recent variants do not appear to cause more severe disease compared with previous variants.[2]
Several recombinant SARS-CoV-2 variants have been identified over the course of the pandemic, and the vast majority do not appear to confer any advantage to the virus.
Currently circulating variants
There are currently no circulating variants of concern, but there are variants of interest and variants under monitoring. Consult your local public health authority for the most current information on circulating variants in your area. The following resources are available:
Use of this content is subject to our disclaimer

