Aetiology
Orthoebolaviruses belong to the Filoviridae family (genus Orthoebolavirus; order: Mononegavirales). These viruses are elongated, filamentous structures of variable length.[Figure caption and citation for the preceding image starts]: Transmission electron micrograph showing some of the ultrastructural morphology displayed by an Ebola virus virionCenters for Disease Control and Prevention [Citation ends].
The virus is thought to be initially acquired from exposure to body fluids or tissue from infected animals such as bats and nonhuman primates; however, the natural reservoir and mode of transmission to humans has not been confirmed.[39] Laboratory testing of reservoir competence shows that successful infection is possible in bats and rodents, but not in plants or arthropods.[40][41][42] Animal-to-human transmission may occur during hunting and consumption of the reservoir species or infected nonhuman primates. The local practice of eating bush meat or food contaminated with bat feces (3 species of tree-roosting bats have been implicated as a reservoir) is also thought to contribute.
[Figure caption and citation for the preceding image starts]: Ebolavirus ecology showing enzootic and epizootic cyclesCenters for Disease Control and Prevention [Citation ends].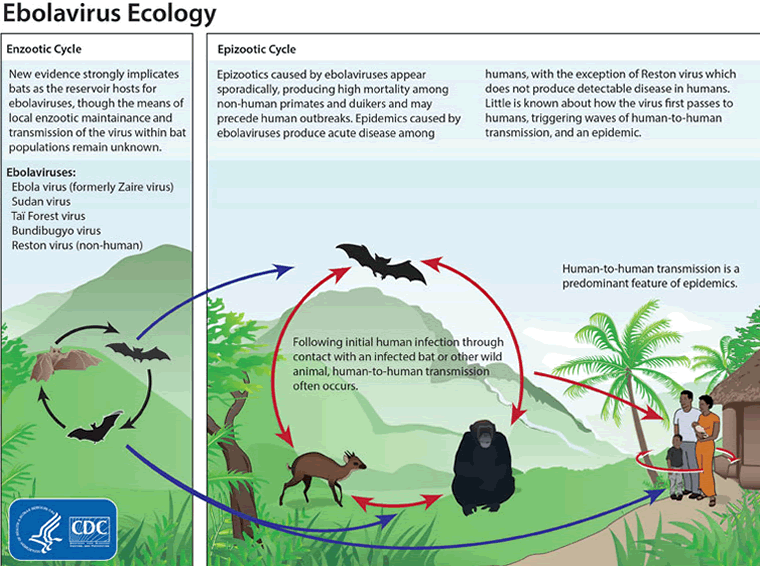
Human-to-human transmission occurs via direct contact with body fluids from infected patients or objects contaminated with infected body fluids.[43][44] In the early epidemics, the reuse of nonsterile injections was responsible for many healthcare-associated transmissions.[24] However, while this still remains a risk, most cases result from close physical contact or contact with body fluids (e.g., sweat, blood, feces, vomit, saliva, genital secretions [including semen], amniotic fluid, and breast milk) of infected patients.
The level of virus in the blood increases during the course of illness and patients are most infectious in the later stages of the disease (i.e., during diarrhea, vomiting, and hemorrhage).[45] Large amounts of virus can be found in the skin and, as sweat may also contain the virus, touching an infected patient may result in transmission.[46]
Super-spreading events in the community are also increasingly recognized as a contributing factor; a funeral of a traditional healer in Sierra Leone in 2015 was linked to 300 cases.[47] In one study, it was found that super-spreaders were responsible for approximately 61% of infections in the 2014 outbreak.[48]
In a study to identify the rate of viral shedding in various body fluids, the virus was isolated from saliva, breast milk, stool, tears, and semen up to 40 days after the onset of illness.[44][49][50] The virus can still be detected in semen more than 12 months after recovery from infection, possibly due to testicular tissue being an immunologically protected site.[51] This means that sexual transmission may be possible long after the infection has resolved, and such cases were confirmed during and following the 2014 outbreak.[44][49][50][52][53][54][55][56] The virus has also been detected in vaginal fluid.[57] Other immunologically protected sites include the interior of the eyes, placenta, and central nervous system, particularly cerebrospinal fluid.[58] Viral shedding may continue from urine and sweat. In one recovered patient in Germany, virus was detected in urine 14 days after it was not detected in serum, and in sweat for up to 19 days after it was not detected in serum.[46]
Infection via the inhalation route has been shown to be possible in nonhuman primates; however, there is no evidence for airborne transmission in humans.[14][59] The possibility of opportunistic airborne transmission of the virus during forceful vomiting (similar to that seen with norovirus infection), and during aerosol-generating procedures associated with critical care interventions, should still be considered.
Outside the endemic areas, Ebola disease is rare and is usually an imported infection.[60] Travelers arriving from affected areas, as well as laboratory scientists and others working with potentially infected materials and animals, are at high risk.
Fisiopatologia
There have been major advances in elucidating the pathogenesis of infection; however, most of the studies have been performed in nonhuman primate and rodent models.[7] This is because of the difficulties in conducting human studies in poorly-resourced settings where these infections naturally occur.
The virus genome consists of a single 19 kb strand of negative-sense RNA with 7 viral genes that are transcribed by the viral RNA-dependent RNA polymerase present in the virion. The single strand of RNA is covered by helically-arranged viral nucleoproteins NP and VP30 that are linked by matrix proteins VP24 and VP4 to the lipid bilayer that coats the virion.[14] There was rapid mutation of the virus in the 2014 outbreak, raising concerns about its ability to evade host immune responses and evolve under pressure of novel therapies.[61][62]
The incubation period after infection is 2 to 21 days.[3] The basic reproduction number (R₀), the number of secondary cases generated by a primary case in a totally susceptible population, has been estimated to be 1.3 to 2. However, R₀ varies significantly across countries and outbreaks. The overall pooled mean R₀ has been estimated to be 1.95, but has been as high as 10 in some outbreaks. In countries where cases are imported, the value ranged from 1.5 in the US, to 1.6 in the UK, and 2.4 in Gambia.[63]
Tissue invasion occurs via infected fluid coming into contact with breaks in the mucosa or skin. This can occur with animal-to-human or human-to-human transmission. Monocytes, macrophages, and dendritic cells are the preferred replication sites for filoviruses on initial infection. Infected cells migrate to the regional lymph nodes, liver, and spleen, thereby disseminating the infection. Ebola virus has a wide cell tropism and is able to infect a variety of different cell types, but extensive viral replication occurs in lymphoid tissue, liver, and the spleen.[7][14][64] It also has the remarkable ability to modulate the expression of genes involved in the host immune response, causing lymphocyte apoptosis and attenuation of the protective effects of interferon.[65][66][67][68][69]
The host immune response is crucial and dictates the outcome of infection. Progression to the severe end of the disease spectrum occurs when the virus triggers expression of a host of pro-inflammatory cytokines, including: interferons; interleukins (IL) such as IL-2, IL- 6, IL-8, and IL-10; interferon inducible protein; and tumor necrosis factor (TNF)-alpha.[7][14][70] This, in turn, causes endothelial activation and reduced vascular integrity, release of tissue factor (with associated onset of coagulopathy), and increased nitric oxide levels (with associated hypotension).[71] Infection leads to lymphocyte depletion through indirect apoptosis (since the virus does not replicate in lymphocytes), and neutrophil suppression via glycoprotein GP.[72] The most common cause of thrombocytopenia is platelet disappearance from damaged tissue or more generalized virus-induced disseminated intravascular coagulation, where coagulation factors are depleted.[73] Disseminated intravascular coagulation, along with acute hepatic impairment, predisposes the patient to bleeding complications. Other complications of severe disease include acute kidney injury, hepatitis, and pancreatitis.[14] Early antibody response, along with reduced lymphocyte depletion, is associated with effective viral clearance and survival.[74] Flow cytometry, which was used in a treatment center in Guinea during the 2014 outbreak, demonstrated that T-cell dysregulation (characterized by higher expression of CTLA-4 and PD-1 on CD4 and CD8 cells) was associated with death. This confirms earlier suggestions that an adequate, but controlled, immune response is key to survival.[75]
The development of shock is still not well understood. Multiple factors may contribute, including: bacterial sepsis, possibly through gut translocation of bacteria; a direct effect of the virus; disseminated intravascular coagulation; or hemorrhage.[70]
Classification
Virus taxonomy
Orthoebolaviruses belong to the Filoviridae family (genus Orthoebolavirus).[1]
There are currently six known species of Orthoebolavirus (note: species names were updated in April 2023):[2]
Ebola virus (EBOV) - Orthoebolavirus zairense (formerly known as Zaire ebolavirus)
Sudan virus (SUDV) - Orthoebolavirus sudanense (formerly known as Sudan ebolavirus)
Tai Forest virus (TAFV) - Orthoebolavirus taiense (formerly known as Tai Forest ebolavirus or Cote d'Ivoire ebolavirus)
Bundibugyo virus (BDBV) - Orthoebolavirus bundibugyoense (formerly known as Bundibugyo ebolavirus)
Reston virus (RESTV) - Orthoebolavirus restonense (formerly known as Reston ebolavirus)
Bombali virus (BOMV) - Orthoebolavirus bombaliense (formerly known as Bombali ebolavirus).
Reston virus and Bombali virus are not known to cause illness in humans.[1]
The four species that cause illness in humans cause slightly different clinical syndromes of varying severity, and have a reported case fatality rate of 25% to 90% across different outbreaks (the average rate is approximately 50%).[3] The Ebola virus and Sudan virus are especially known for their virulence; the other species are considered to be less virulent.
Ebola virus (Orthoebolavirus zairense):
First isolated in 1976 during an outbreak in northern Zaire (now known as the Democratic Republic of the Congo [DRC]).[1] Seems to be the most virulent of the six species and has the highest case fatality rate out of all species.[4] It is responsible for the largest outbreak that started in West Africa in 2014.
Sudan virus (Orthoebolavirus sudanense):
First isolated in 1976 during an outbreak in southern Sudan. Causes an identical syndrome to Orthoebolavirus zairense; however, the case fatality rate is lower.[4]
Tai Forest virus (Orthoebolavirus taiense):
Only one case has been documented in 1994 in a Swiss researcher who performed an autopsy on a dead chimpanzee in Tai National Park in Cote d’Ivoire.[5] She recovered from the febrile phase of the illness with no hemorrhagic complications.
Bundibugyo virus (Orthoebolavirus bundibugyoense):
Discovered in 2007 during a single outbreak in the Bundibugyo district of western Uganda. The isolated virus was identified as a distinct species, distantly related to Orthoebolavirus taiense.[6]
Reston virus (Orthoebolavirus restonense):
First isolated in Reston, Virginia, US in 1989 where it was found in Cynomolgus monkeys imported from the Philippines. Several workers exposed to infected animals were found to have positive serology, but no clinical symptoms. Since then, the virus has also been isolated from swine in the Philippines.[7][8]
Bombali virus (Orthoebolavirus bombaliense):
First discovered in Sierra Leone in 2018 in the organs of the Angolan free-tailed bat (Mops condylurus) and the little free-tailed bat (Chaerephon pumilus). It has also been identified in bats in Kenya and Guinea.[9][10] It is unknown whether this virus is pathogenic in humans.[11]
Other filoviral infections
The Filoviridae family of viruses includes orthoebolaviruses, orthomarburgviruses, and Cuevavirus, among others. Orthomarburgviruses are the only other member of this group known to cause human infection. It has been isolated from bats and causes a similar syndrome to Ebola disease. Several outbreaks have been reported, often related to animal exposure in mines or caves.[2][12]
Use of this content is subject to our disclaimer
